RNA Barcoding
Betainfluenzavirus
Influenza B virus
Sequence Information
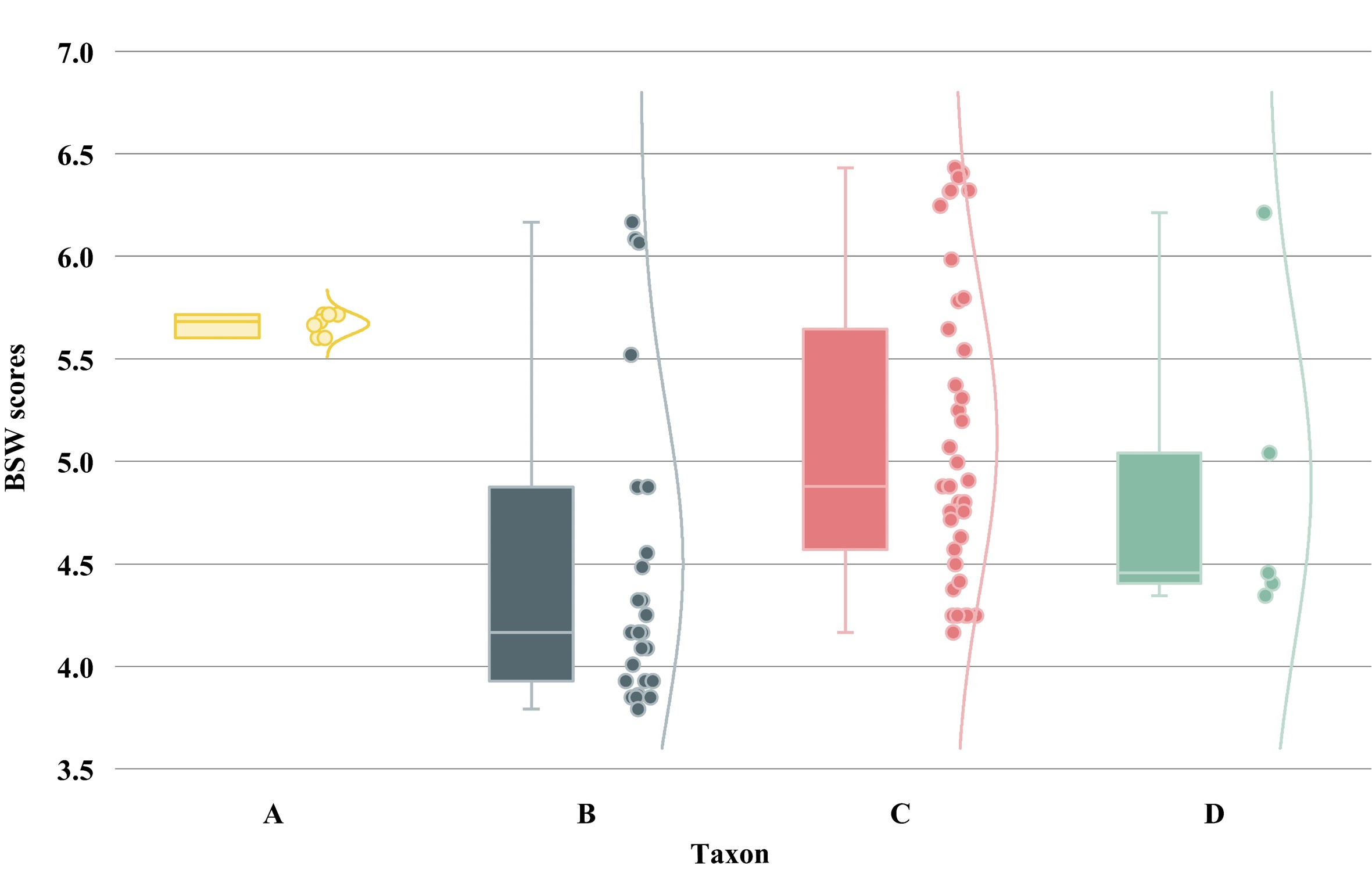
| Serial numbers | Length | P value | Max total score | Max E value | BSW score | Regions |
|---|---|---|---|---|---|---|
| 1 | 35 | 0.0045 | 65.8 | 9.00E-08 | 4.17 | PB2 |
|
>Barcode_1
AGAATATCAGGAAGAGGATTCAAAAATGATGAAGA Forward primer AGAATATCAGGAAGAGGAT Reverse primer TCTTCATCATTTTTGAATC |
||||||
| 2 | 35 | 0.0045 | 65.8 | 9.00E-08 | 4.17 | PB2 |
|
>Barcode_2
GACATGTTCCAATGGGATGCATTTGAAGCATTTGA Forward primer GACATGTTCCAATGGGATG Reverse primer TCAAATGCTTCAAATGCA |
||||||
| 3 | 34 | 0.0052 | 63.9 | 3.00E-07 | 4.09 | PB2 |
|
>Barcode_3
TAACTATATGCGGCAGAATGATGTCATTAAAAGG Forward primer TAACTATATGCGGCAGAA Reverse primer CCTTTTAATGACATCATTC |
||||||
| 4 | 31 | 0.0083 | 58.4 | 1.00E-05 | 3.85 | PB1 |
|
>Barcode_4
CAGTAAAGAATATAAAGAAAAAATTGCCTGC Forward primer CAGTAAAGAATATAAAGAA Reverse primer GCAGGCAATTTTT |
||||||
| 5 | 32 | 0.0071 | 60.2 | 3.00E-06 | 3.93 | PB1 |
|
>Barcode_5
TTTGTATTAGTAGTTGAAAACTTGGCTAAAAA Forward primer TTTGTATTAGTAGTTGAAA Reverse primer TTTTTAGCCAAGTTTTCAA |
||||||
| 6 | 32 | 0.0071 | 60.2 | 3.00E-06 | 3.93 | PB1 |
|
>Barcode_6
ATGATGATGGGAATGTTTAATATGCTATCTAC Forward primer ATGATGATGGGAATGTTT Reverse primer GTAGATAGCATATTAAACA |
||||||
| 7 | 40 | 0.0021 | 75.0 | 3.00E-10 | 4.55 | PB1 |
|
>Barcode_7
TATTGGGAATAAACATGAGCAAAAAGAAAAGTTACTGTAA Forward primer TATTGGGAATAAACATGAG Reverse primer TTACAGTAACTTTTCTTTT |
||||||
| 8 | 65 | 0.0001 | 121 | 2.00E-23 | 6.08 | PA |
|
>Barcode_8
TTCCAGACTACAATAATACAAAAGGCCAAAAACACAATGGCAGAATTTAGTGAAGATCCTGAATT Forward primer TTCCAGACTACAATAATAC Reverse primer AATTCAGGATCTTCACT |
||||||
| 9 | 35 | 0.0045 | 65.8 | 9.00E-08 | 4.17 | PA |
|
>Barcode_9
GGATTGGCTGATGATTACTTTTGGAAAAAGAAAGA Forward primer GGATTGGCTGATGATTACT Reverse primer TCTTTCTTTTTCCAAAAGT |
||||||
| 10 | 37 | 0.0033 | 69.4 | 1.00E-08 | 4.32 | PA |
|
>Barcode_10
ACCATGAGCTACCAGAAGTTCCATATAATGCCTTTCT Forward primer ACCATGAGCTACCAGAAGT Reverse primer AGAAAGGCATTATATGGAA |
||||||
| 11 | 35 | 0.0045 | 65.8 | 9.00E-08 | 4.17 | PA |
|
>Barcode_11
AGAGTGGACTCAGGAAAGTGGCCAAAATATACTGT Forward primer AGAGTGGACTCAGGAAAGT Reverse primer ACAGTATATTTTGGCCACT |
||||||
| 12 | 36 | 0.0038 | 67.6 | 3.00E-08 | 4.25 | HA |
|
>Barcode_12
AAAATGAAGGCAATAATTGTACTACTCATGGTAGTA Forward primer AAAATGAAGGCAATAATTG Reverse primer TACTACCATGAGTA |
||||||
| 13 | 44 | 0.0011 | 82.4 | 3.00E-12 | 4.87 | HA |
|
>Barcode_13
GGAGGATGGGAAGGAATGATTGCAGGTTGGCACGGATACACATC Forward primer GGAGGATGGGAAGGAATGA Reverse primer GATGTGTATCCGTGCCAAC |
||||||
| 14 | 37 | 0.0033 | 69.4 | 1.00E-08 | 4.32 | NP |
|
>Barcode_14
TAAAAGAACTGAAAATCAAAATGTCCAACATGGATAT Forward primer TAAAAGAACTGAAAATCAA Reverse primer ATATCCATGTTGG |
||||||
| 15 | 39 | 0.0024 | 73.1 | 1.00E-09 | 4.48 | NP |
|
>Barcode_15
AATTACCTTTTTAAAAGAAGAGGTGAAAACAATGTACAA Forward primer AATTACCTTT Reverse primer TTGTACATTGTTTTCACCT |
||||||
| 16 | 44 | 0.0011 | 82.4 | 3.00E-12 | 4.87 | NP |
|
>Barcode_16
AGACCTATTGCTCTAAGCAAGCAAGCTGTAAGAAGAATGCTGTC Forward primer AGACCTATTGCTCTAAGC Reverse primer GACAGCATTCTTCTTACAG |
||||||
| 17 | 32 | 0.0071 | 60.2 | 3.00E-06 | 3.93 | NP |
|
>Barcode_17
CGTGATGCAGATGTCAAAGGAAATCTACTCAA Forward primer CGTGATGCAGATGTCAAAG Reverse primer TTGAGTAGATTTCCTTTGA |
||||||
| 18 | 63 | 0.0001 | 117 | 2.00E-22 | 6.07 | NP |
|
>Barcode_18
AAGAAAACCAGTGGAAATGCTTTCATTGGGAAGAAAATGTTTCAAATATCAGACAAAAACAAA Forward primer AAGAAAACCAGTGGAAATG Reverse primer TTTGTTTTTGTCTGATATT |
||||||
| 19 | 79 | 0.0001 | 147 | 4.00E-31 | 6.17 | NP |
|
>Barcode_19
ATTCCAATTAAGCAGACCATCCCCAATTTCTTCTTTGGGAGGGACACAGCAGAGGATTATGATGACCTCGATTATTAAA Forward primer ATTCCAATTAAGCAGACCA Reverse primer TTTAATAATCGAGGTCATC |
||||||
| 20 | 32 | 0.0083 | 60.2 | 3.00E-06 | 3.86 | NA |
|
>Barcode_20
AGGCCAAAAAATGAACAATGCTACCTTCAACT Forward primer AGGCCAAAAA Reverse primer AGTTGAAGGTAGCATTGTT |
||||||
| 21 | 31 | 0.0083 | 58.4 | 1.00E-05 | 3.85 | NA |
|
>Barcode_21
TAAAATATGGAGAAGCATATACTGACACATA Forward primer TAAAATATGGAGAAGCAT Reverse primer TATGTGTCAGTATATGCTT |
||||||
| 22 | 31 | 0.0083 | 58.4 | 1.00E-05 | 3.85 | M |
|
>Barcode_22
AATGTCGCTGTTTGGAGACACAATTGCCTAC Forward primer AATGTCGCTGTTTGGAGAC Reverse primer GTAGGCAATTGTGTCTCCA |
||||||
| 23 | 30 | 0.0097 | 60.0 | 1.00E-05 | 3.79 | M |
|
>Barcode_23
AGAAGATGGAGAAGGCAAAGCAGAACTAGC Forward primer AGAAGATGGAGAAGGCAAA Reverse primer GCTAGTTCTGCTTTGCCT |
||||||
| 24 | 35 | 0.0045 | 65.8 | 9.00E-08 | 4.17 | M |
|
>Barcode_24
TTATCAGCTCTCCATTTCATGGCTTGGACAATAGG Forward primer TTATCAGCTCTCCATTTCA Reverse primer CCTATTGTCCAAGCCATGA |
||||||
| 25 | 34 | 0.0052 | 63.9 | 3.00E-07 | 4.09 | M |
|
>Barcode_25
ACTGTATTTCTTACTATGCATTTAAGCAAATTGT Forward primer ACTGTATTTCTTACTATGC Reverse primer ACAATTTGCTTAAATGCA |
||||||
| 26 | 44 | 0.0011 | 82.4 | 3.00E-12 | 4.87 | NS |
|
>Barcode_26
ACCACAACACAAATTGAGGTGGGTCCGGGAGCAACCAATGCCAC Forward primer ACCACAACACAAATTGAGG Reverse primer GTGGCATTGGTTG |
||||||
| 27 | 33 | 0.0061 | 62.1 | 9.00E-07 | 4.01 | NS |
|
>Barcode_27
TCTGCTGGAATTGAAGGGTTTGAGCCATACTGT Forward primer TCTGCTGGAATTGAAGGGT Reverse primer ACAGTATGGCTCAAACCCT |
||||||
| 28 | 63 | 0.0001 | 117 | 2.00E-22 | 6.07 | NS |
|
>Barcode_28
TGGGAGTCTTATCCCAATTTGGTCAAGAGCACCGATTATCACCAGAAGAGGGAGACAATTAGA Forward primer TGGGAGTCTTATCC Reverse primer TCTAATTGTCTCCCTCTTC |
||||||
| 29 | 53 | 0.0003 | 99 | 5.00E-17 | 5.52 | NS |
|
>Barcode_29
GAAACATTGTATGAAATGAAGGATGTGGTTGAAGTGTACAGCAGGCAGTGCTT Forward primer GAAACATTGTATGAAATGA Reverse primer AAGCACTGCCTG |
||||||